swanlab.Molecule
| Parameter | Description |
|---|---|
| pdb_data | (str) PDB data received (in string format). |
| caption | (str) Label for the molecule object. Used to tag the molecule when displayed in the experiment dashboard. |
Overview
Converts various types of biochemical molecules for recording via swanlab.log().
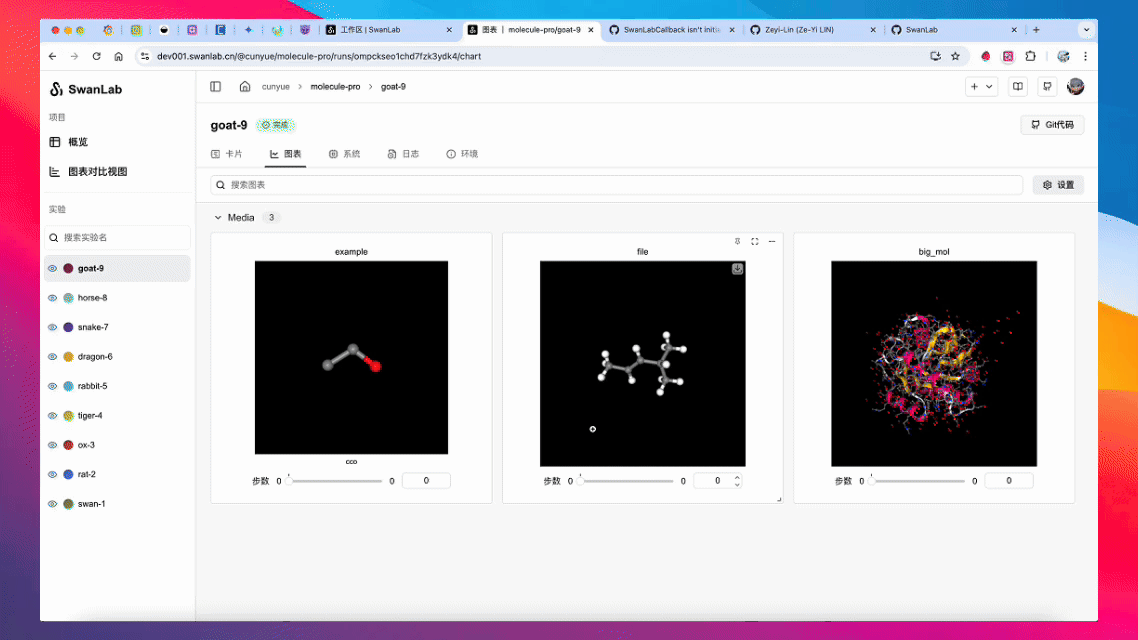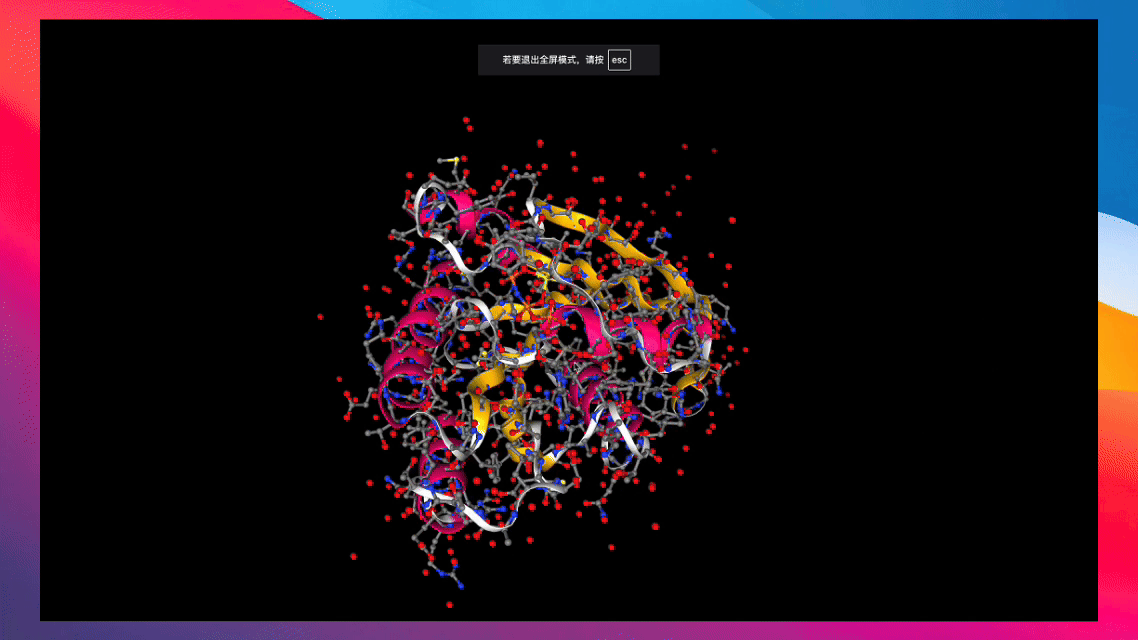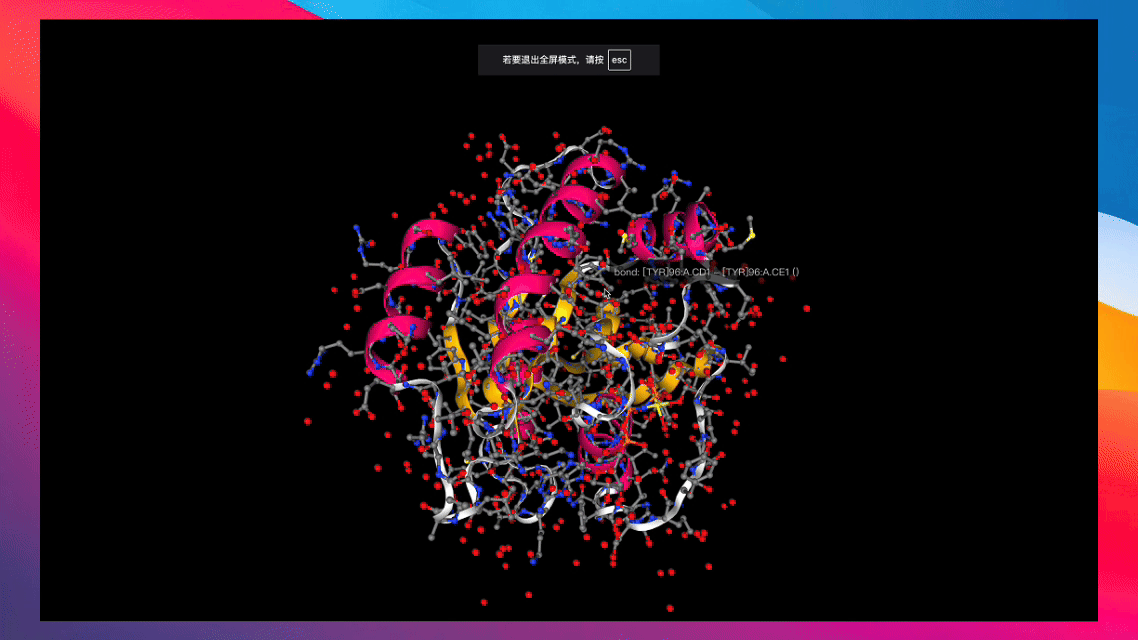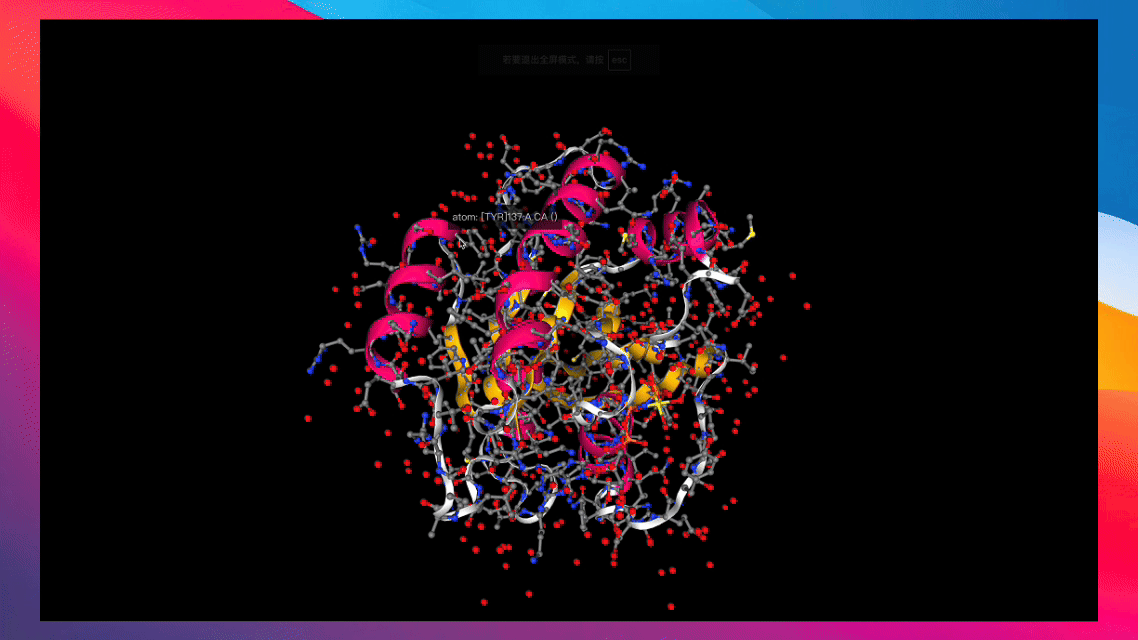
Create from RDKit Mol Object
python
from rdkit import Chem
import swanlab
mol = Chem.MolFromSmiles("CCO")
molecule = swanlab.Molecule.from_mol(mol, caption="Ethanol")
swanlab.init(project="molecule_demo")
swanlab.log({"molecule": molecule})Create from PDB File
python
import swanlab
molecule = swanlab.Molecule.from_pdb("path/to/your/pdb/file.pdb")
swanlab.init(project="molecule_demo")
swanlab.log({"molecule": molecule})Create from SDF File
python
import swanlab
molecule = swanlab.Molecule.from_sdf("path/to/your/sdf/file.sdf")
swanlab.init(project="molecule_demo")
swanlab.log({"molecule": molecule})Create from SMILES String
python
import swanlab
molecule = swanlab.Molecule.from_smiles("CCO")
swanlab.init(project="molecule_demo")
swanlab.log({"molecule": molecule})Create from MOL File
python
import swanlab
molecule = swanlab.Molecule.from_mol("path/to/your/mol/file.mol")
swanlab.init(project="molecule_demo")
swanlab.log({"molecule": molecule})